PCA Features and Analysis
When dealing with substitution effects in your data, a practical approach involves the orthogonalization of features—this can be achieved by applying Principal Component Analysis (PCA). You may know PCA as a technique to reduce the dimensionality of a data set, but it can also be a useful tool to verify the patterns recognized by feature importance.
Imagine you use PCA to create orthogonal features. This unsupervised learning method ranks your features based on their statistical significance, without any reference to the labels of your data set. The advantage here is that PCA’s assessment of feature importance is possibly less susceptible to overfitting due to it being an unsupervised learning algorithm.
Note
Unsupervised machine learning algorithms are still susceptible to overfitting. However, the way PCA operates (i.e. it transforms the dataset based on the variance of the features) means it is largely exempt of this concern.
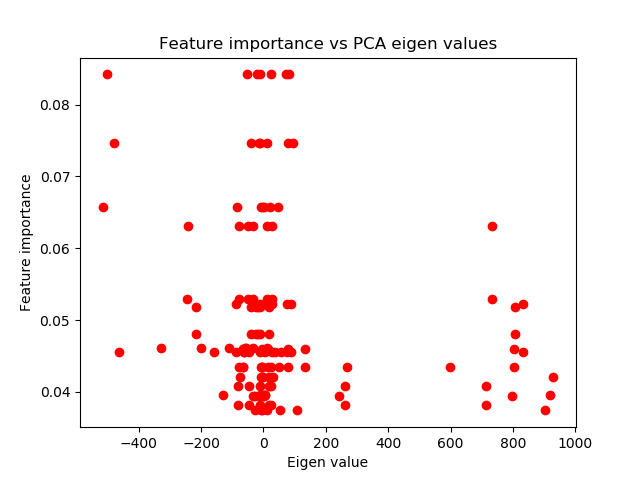
Let’s consider an analysis using Mean Decrease Impurity (MDI), Mean Decrease Accuracy (MDA), or Single Feature Importance (SFI) methods. These techniques do consider label information when determining the most important features. If the features selected as most important by MDI, MDA, or SFI are the same ones that PCA deemed “principal”, then this provides supportive evidence. Specifically, it suggests that your machine learning algorithm’s identified patterns are likely to be robust and not the result of overfitting.
Implementation
Module which implements feature PCA compression and PCA analysis of feature importance.
- feature_pca_analysis(feature_df, feature_importance, variance_thresh=0.95)
-
Perform correlation analysis between feature importance (MDI for example, supervised) and PCA eigenvalues (unsupervised).
High correlation means that probably the pattern identified by the ML algorithm is not entirely overfit.
- Parameters:
-
-
feature_df – (pd.DataFrame): Features dataframe.
-
feature_importance – (pd.DataFrame): Individual MDI feature importance.
-
variance_thresh – (float): Percentage % of overall variance which compressed vectors should explain in PCA compression.
-
- Returns:
-
(dict): Dictionary with kendall, spearman, pearson and weighted_kendall correlations and p_values.
- get_orthogonal_features(feature_df, variance_thresh=0.95, num_features=None)
-
Advances in Financial Machine Learning, Snippet 8.5, page 119.
Computation of Orthogonal Features.
Gets PCA orthogonal features.
- Parameters:
-
-
feature_df – (pd.DataFrame): Dataframe of features.
-
variance_thresh – (float): Percentage % of overall variance which compressed vectors should explain. Default is 0.95.
-
num_features – (int) Manually set number of features, overrides variance_thresh. Default is None.
-
- Returns:
-
(np.array): Compressed PCA features which explain %variance_thresh of variance.
- get_pca_rank_weighted_kendall_tau(feature_imp, pca_rank)
-
Advances in Financial Machine Learning, Snippet 8.6, page 121.
Computes Weighted Kendall’s Tau Between Feature Importance and Inverse PCA Ranking.
- Parameters:
-
-
feature_imp – (np.array): Feature mean importance.
-
pca_rank – (np.array): PCA based feature importance rank.
-
- Returns:
-
(float): Weighted Kendall Tau of feature importance and inverse PCA rank with p_value.
Example
>>> from sklearn.tree import DecisionTreeClassifier
>>> from sklearn.ensemble import BaggingClassifier
>>> from mlfinlab.util.generate_dataset import generate_classification_dataset
>>> from mlfinlab.feature_importance.importance import mean_decrease_impurity
>>> from mlfinlab.feature_importance.orthogonal import (
... get_orthogonal_features,
... feature_pca_analysis,
... )
>>> # Generate toy dataset
>>> X, y = generate_classification_dataset(n_features=50, n_samples=1000)
>>> # Create, fit and cross-validate the model
>>> base_estimator = DecisionTreeClassifier(
... class_weight="balanced",
... random_state=42,
... max_depth=3,
... criterion="entropy",
... min_samples_leaf=4,
... min_samples_split=3,
... max_features="auto",
... )
>>> clf = BaggingClassifier(
... n_estimators=452,
... n_jobs=-1,
... random_state=42,
... oob_score=True,
... base_estimator=base_estimator,
... )
>>> clf = clf.fit(X, y)
>>> # Produce the MDI feature importance map
>>> mdi_feature_imp = mean_decrease_impurity(clf, X.columns)
>>> # Use PCA to get orthogonal features and do analysis
>>> get_orthogonal_features(X)
array(...)
>>> feature_pca_analysis(X, mdi_feature_imp)
{'Pearson': ..., 'Spearman': ..., 'Kendall': ..., 'Weighted_Kendall_Rank': ...}