Note
The following implementation and documentation closely follow the work of Donnat, P., Marti, G. and Very, P., Toward a generic representation of random variables for machine learning.
Correlated Random Walks
Being able to discriminate random variables both on distribution and dependence on time series is motivated by the study of financial assets returns. The authors proposed a distance metric (GNPR) that “improves the performance of machine learning algorithms working on independent and identically distributed stochastic processes”.
As examined by the authors, there is a need for a generic representation of random variables for machine learning. They introduce a non-parametric approach to represent random variables that is able to split and detect different underlying distributions on a time series. This method is called the generic non-parametric representation (GNPR) approach, the authors have shown it separates distributions more effectively than other methods such as generic parametric representation (GPR), \(L_2\) distance, and distance correlation.
Note
The GNPR approach is described in our documentation, located in the Codependence by Marti section.
Note
Underlying Literature
The following sources elaborate extensively on the topic:
-
Toward a generic representation of random variables for machine learning by Donnat, P., Marti, G. and Very, P.
Time Series Generation with Different Distributions
In order to test and verify the efficiency of this approach, the authors provide a method to generate time series datasets. They are defined as \(N\) time series, each of length \(T\), which are subdivided into \(K\) correlation clusters, themselves subdivided into \(D\) distribution clusters.
If \(\textbf{W}\) is sampled from a normal distribution \(N(0, 1)\) of length \(T\), \((Y_k)_{k=1}^K\) is \(K\) i.i.d random distributions each of length \(T\), and \((Z_d^i)_{d=1}^D\); for \(i \leq i \leq N\) are independent random distributions of length \(T\), for \(i \leq i \leq N\) they define:
Where
-
\(\alpha_{d, i} = 1\), if \(i \equiv d - 1\) (mod \(D\)), 0 otherwise
-
\(\beta \in [0, 1]\)
-
\(\beta_{k, i} = \beta\), if \(\textit{ceil}(iK/N) = k\), 0 otherwise.
The authors show that even though the mean and the variance of the \((Y_k)\) and \((Z_d^i)\) distributions are the same and their variables are highly correlated, GNPR is able to successfully separate them into different clusters.
The distributions supported by default are:
-
Normal distribution (
np.random.normal) -
Laplace distribution (
np.random.laplace) -
Student’s t-distribution (
np.random.standard_t)
Implementation
To override the default distributions used to create the time series, the user must pass a list of the names of the distributions to use as the parameter
dists_clusters. The first value of this list is used to generate \((Y_k)_{k=1}^K\). The available distributions are:
-
“normal” (
np.random.normal(0, 1)) -
“normal_2” (
np.random.normal(0, 2)) -
“laplace” (
np.random.laplace(0, 1 / np.sqrt(2))) -
“student-t” (
np.random.standard_t(3) / np.sqrt(3))
- generate_cluster_time_series(n_series, t_samples=100, k_corr_clusters=1, d_dist_clusters=1, rho_main=0.1, rho_corr=0.3, price_start=100.0, dists_clusters=('normal', 'normal', 'student-t', 'normal', 'student-t'))
-
Generates a synthetic time series of correlation and distribution clusters.
It is reproduced with modifications from the following paper: Donnat, P., Marti, G. and Very, P., 2016. Toward a generic representation of random variables for machine learning. Pattern Recognition Letters, 70, pp.24-31.
This method creates n_series time series of length t_samples. Each time series is divided into k_corr_clusters correlation clusters. Each correlation cluster is subdivided into d_dist_clusters distribution clusters. A main distribution is sampled from a normal distribution with mean = 0 and stdev = 1, adjusted by a rho_main factor. The correlation clusters are sampled from a given distribution, are generated once, and adjusted by a rho_corr factor. The distribution clusters are sampled from other given distributions, and adjusted by (1 - rho_main - rho_corr). They are sampled for each time series. These three series are added together to form a time series of returns. The final time series is the cumulative sum of the returns, with a start price given by price_start.
- Parameters:
-
-
n_series – (int) Number of time series to generate.
-
t_samples – (int) Number of samples in each time series.
-
k_corr_clusters – (int) Number of correlation clusters in each time series.
-
d_dist_clusters – (int) Number of distribution clusters in each time series.
-
rho_main – (float): Strength of main time series distribution.
-
rho_corr – (float): Strength of correlation cluster distribution.
-
price_start – (float) Starting price of the time series.
-
dists_clusters – (list) List containing the names of the distributions to sample from. The following numpy distributions are available: “normal” = normal(0, 1), “normal_2” = normal(0, 2), “student-t” = standard_t(3)/sqrt(3), “laplace” = laplace(1/sqrt(2)). The first disitribution is used to sample for the correlation clusters (k_corr_clusters), the remaining ones are used to sample for the distribution clusters (d_dist_clusters).
-
- Returns:
-
(pd.DataFrame) Generated time series. Has size (t_samples, n_series).
Example
The authors provide multiple parameters and distributions in their paper. \(N\) represents the normal distribution, \(L\) represents \(Laplace(0, 1/\sqrt{2})\), and \(S\) represents \(t-distribution(3)/\sqrt{3}\)
Clustering |
N |
T |
K |
D |
|
|
\(Y_k\) |
\(Z_1^i\) |
\(Z_2^i\) |
\(Z_3^i\) |
\(Z_4^i\) |
Distribution |
200 |
5000 |
1 |
4 |
0.1 |
0 |
\(N(0,1)\) |
\(N(0,1)\) |
\(L\) |
\(S\) |
\(N(0,2)\) |
Dependence |
200 |
5000 |
10 |
1 |
0.1 |
0.3 |
\(S\) |
\(S\) |
\(S\) |
\(S\) |
\(S\) |
Mix |
200 |
5000 |
5 |
2 |
0.1 |
0.3 |
\(N(0,1)\) |
\(N(0,1)\) |
\(S\) |
\(N(0,1)\) |
\(S\) |
The Distribution example generates a time series that has a global normal distribution, no correlation clustering, and 4 distribution clusters.

(Top) Time series plot. (Left) GPR codependence matrix. Only two apparent clusters are seen with no indication of a global embedded distribution. (Right). All 4 distributions clusters can be seen, as well as the global embedded distribution.
The Dependence example generates a time series that has a global normal distribution, 10 correlation clusters, and no distribution clusters.
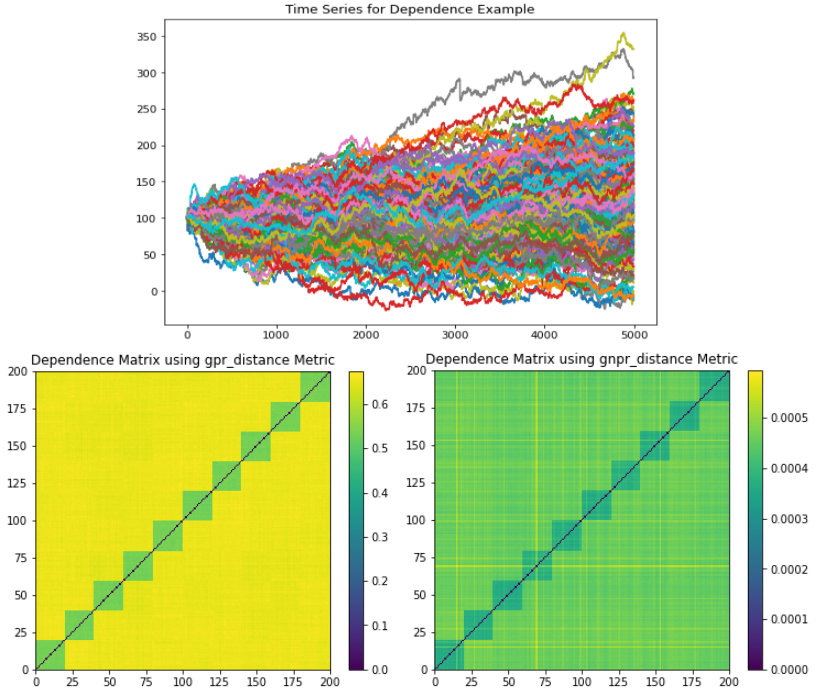
(Top) Time series plot. (Left) GPR codependence matrix. Only 10 correlation clusters are seen with no indication of a global embedded distribution. All 10 correlation clusters can be seen, as well as the global embedded distribution.
The Mix example generates a time series that has a global normal distribution, 5 correlation clusters, and 2 distribution clusters.
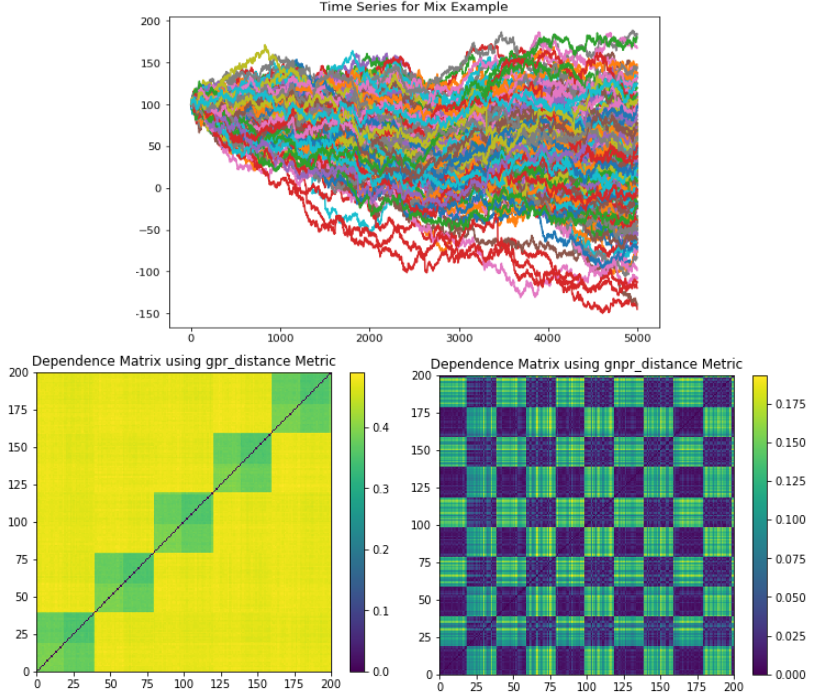
(Top) Time series plot. (Left) GPR codependence matrix. Only 5 correlation clusters are seen with no indication of a global embedded distribution. All 5 correlation clusters and 2 distribution clusters can be seen, as well as the global embedded distribution.
import matplotlib.pyplot as plt
from mlfinlab.data_generation.correlated_random_walks import generate_cluster_time_series
from mlfinlab.data_generation.data_verification import plot_time_series_dependencies
# Initialize the example parameters for the time series
n_series = 200
t_samples = 5000
k_clusters = [1, 10, 5]
d_clusters = [4, 1, 2]
rho_corrs = [0, 0.3, 0.3]
thetas = [0, 1, 0.5]
dists_clusters = [["normal", "normal", "laplace", "student-t", "normal_2"],
["student-t", "student-t", "student-t", "student-t", "student-t"],
["normal", "normal", "student-t", "normal", "student-t"]]
titles = ["Distribution", "Dependence", "Mix"]
# Plot the time series and codependence matrix for each example
for i in range(len(k_clusters)):
dataset = generate_cluster_time_series(n_series=n_series, t_samples=t_samples, k_corr_clusters=k_clusters[i],
d_dist_clusters=d_clusters[i], rho_corr=rho_corrs[i],
dists_clusters=dists_clusters[i])
dataset.plot(legend=None, title="Time Series for {} Example".format(titles[i]))
plt.figure()
plot_time_series_dependencies(dataset, dependence_method='gpr_distance', theta=thetas[i])
plot_time_series_dependencies(dataset, dependence_method='gnpr_distance', theta=thetas[i])
plt.figure()
plt.show()
Research Notebook
The following research notebook can be used to better understand Correlated Random Walks.